Manual
LR Filter is a software application for logistic regression based variant prioritization for whole-genome sequencing. It is applied to variant call files and aims to reduce false positives due to the bias of a sequencing platform including its short read alignment and variant calling pipeline. The probability of a variant being true positive is modeled using logistic regression with genomic context and genotype quality score. A model is fitted using 6 factors: 1) genotype quality score reported by the sequencing platform, 2) reported in dbSNP database, 3) overlap with the RepeatMasker, 4) present in the other family members (parents and children), 5) genic vs. intergenic elements by RefSeq, and 6) SNV substitution type (or INDEL length). A logistic regression model is trained using annotated variant call files with labeled variants. Variants can be labeled using a set of gold standard variants, e.g., variants concordantly called by two or more sequencing platforms and high-confidence benchmark calls that were compiled by the Genome in a Bottle Consortium.
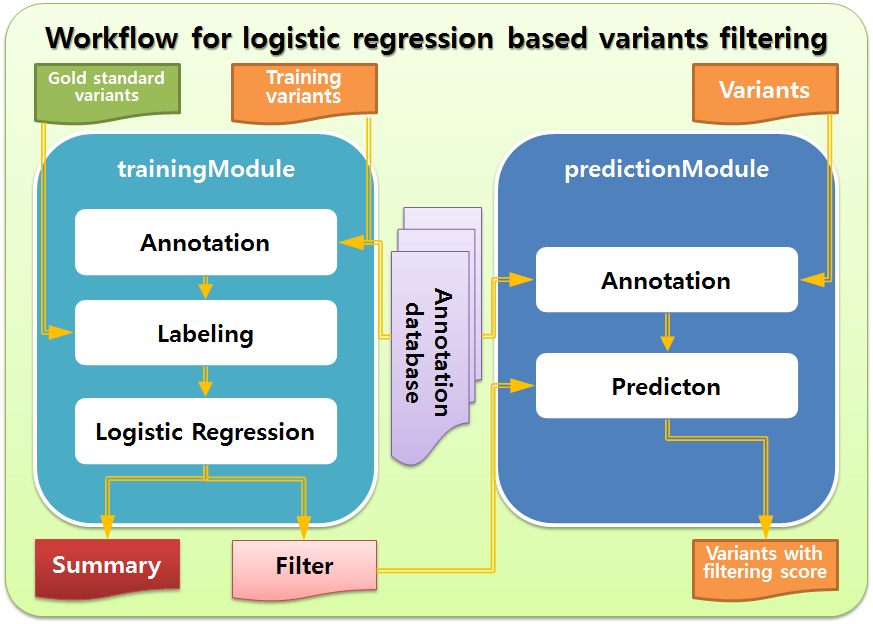
LR Filter consists of two modules: trainingModule and predictionModule.
trainingModule builds logistic regression based filter. It requires training variant call file, annotation databases (dbSNP, RepeatMasker track, RefSeq gene model), and gold standard variant file. It uses gSearch to annotate training variant call file and label variants in it. It also needs R for logistic regression. As output, trainingModule produces logistic regression filter and log files.
predictionModule applies logistic regression based filter to variant call file, assigning the probability of being true positive to each variant. It needs the same annotation databases used in trainingModule and gSearch.
LR Filter is a set of shell scripts, C source code, and python programs. All these scripts, source code, programs, annotation database files, and gSearch should be in the same directory.
Parameters of trainingModule
| Name | Usage | Description |
|---|---|---|
| Training variant call file | -i <training_variant_call_file.gvf> |
A list of training variants in Genome Variation Format (GVF) used for logistic regression |
| Gold standard variant file | -r <gold_standard_variant.gvf> |
A list of gold standard variants in GVF used for labeling training variants |
Parameters of predictionModule
| Name | Usage | Description |
|---|---|---|
| Input variant call file | -i <input_variant_call_file.gvf> |
A list of variants in GVF |
| Logistic regression filter | -f <logistic_regression_filter> |
A logistic regression filter built by trainingModule |
Example of Usage
./trainingModule.sh -i 12877_ill_hg19.gvf -r 12877_ill_hg19_gs.gvf
./predictionModule.sh -i 12877_ill_hg19.gvf -f 12877_ill.gvf.filter